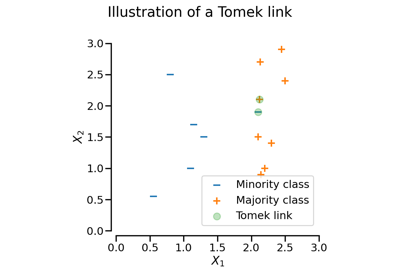
TomekLinks#
- class imblearn.under_sampling.TomekLinks(*, sampling_strategy='auto', n_jobs=None)[source]#
Under-sampling by removing Tomek’s links.
Read more in the User Guide.
- Parameters:
- sampling_strategystr, list or callable
Sampling information to sample the data set.
When
str, specify the class targeted by the resampling. Note the the number of samples will not be equal in each. Possible choices are:'majority': resample only the majority class;'not minority': resample all classes but the minority class;'not majority': resample all classes but the majority class;'all': resample all classes;'auto': equivalent to'not minority'.When
list, the list contains the classes targeted by the resampling.When callable, function taking
yand returns adict. The keys correspond to the targeted classes. The values correspond to the desired number of samples for each class.
- n_jobsint, default=None
Number of CPU cores used during the cross-validation loop.
Nonemeans 1 unless in ajoblib.parallel_backendcontext.-1means using all processors. See Glossary for more details.
- Attributes:
- sampling_strategy_dict
Dictionary containing the information to sample the dataset. The keys corresponds to the class labels from which to sample and the values are the number of samples to sample.
- sample_indices_ndarray of shape (n_new_samples,)
Indices of the samples selected.
Added in version 0.4.
- n_features_in_int
Number of features in the input dataset.
Added in version 0.9.
- feature_names_in_ndarray of shape (
n_features_in_,) Names of features seen during
fit. Defined only whenXhas feature names that are all strings.Added in version 0.10.
See also
EditedNearestNeighboursUndersample by samples edition.
CondensedNearestNeighbourUndersample by samples condensation.
RandomUnderSamplerRandomly under-sample the dataset.
Notes
This method is based on [1].
Supports multi-class resampling. A one-vs.-rest scheme is used as originally proposed in [1].
References
Examples
>>> from collections import Counter >>> from sklearn.datasets import make_classification >>> from imblearn.under_sampling import TomekLinks >>> X, y = make_classification(n_classes=2, class_sep=2, ... weights=[0.1, 0.9], n_informative=3, n_redundant=1, flip_y=0, ... n_features=20, n_clusters_per_class=1, n_samples=1000, random_state=10) >>> print('Original dataset shape %s' % Counter(y)) Original dataset shape Counter({1: 900, 0: 100}) >>> tl = TomekLinks() >>> X_res, y_res = tl.fit_resample(X, y) >>> print('Resampled dataset shape %s' % Counter(y_res)) Resampled dataset shape Counter({1: 897, 0: 100})
Methods
fit(X, y, **params)Check inputs and statistics of the sampler.
fit_resample(X, y, **params)Resample the dataset.
get_feature_names_out([input_features])Get output feature names for transformation.
Get metadata routing of this object.
get_params([deep])Get parameters for this estimator.
is_tomek(y, nn_index, class_type)Detect if samples are Tomek's link.
set_params(**params)Set the parameters of this estimator.
- fit(X, y, **params)[source]#
Check inputs and statistics of the sampler.
You should use
fit_resamplein all cases.- Parameters:
- X{array-like, dataframe, sparse matrix} of shape (n_samples, n_features)
Data array.
- yarray-like of shape (n_samples,)
Target array.
- Returns:
- selfobject
Return the instance itself.
- fit_resample(X, y, **params)[source]#
Resample the dataset.
- Parameters:
- X{array-like, dataframe, sparse matrix} of shape (n_samples, n_features)
Matrix containing the data which have to be sampled.
- yarray-like of shape (n_samples,)
Corresponding label for each sample in X.
- Returns:
- X_resampled{array-like, dataframe, sparse matrix} of shape (n_samples_new, n_features)
The array containing the resampled data.
- y_resampledarray-like of shape (n_samples_new,)
The corresponding label of
X_resampled.
- get_feature_names_out(input_features=None)[source]#
Get output feature names for transformation.
- Parameters:
- input_featuresarray-like of str or None, default=None
Input features.
If
input_featuresisNone, thenfeature_names_in_is used as feature names in. Iffeature_names_in_is not defined, then the following input feature names are generated:["x0", "x1", ..., "x(n_features_in_ - 1)"].If
input_featuresis an array-like, theninput_featuresmust matchfeature_names_in_iffeature_names_in_is defined.
- Returns:
- feature_names_outndarray of str objects
Same as input features.
- get_metadata_routing()[source]#
Get metadata routing of this object.
Please check User Guide on how the routing mechanism works.
- Returns:
- routingMetadataRequest
A
MetadataRequestencapsulating routing information.
- get_params(deep=True)[source]#
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- static is_tomek(y, nn_index, class_type)[source]#
Detect if samples are Tomek’s link.
More precisely, it uses the target vector and the first neighbour of every sample point and looks for Tomek pairs. Returning a boolean vector with True for majority Tomek links.
- Parameters:
- yndarray of shape (n_samples,)
Target vector of the data set, necessary to keep track of whether a sample belongs to minority or not.
- nn_indexndarray of shape (len(y),)
The index of the closes nearest neighbour to a sample point.
- class_typeint or str
The label of the minority class.
- Returns:
- is_tomekndarray of shape (len(y), )
Boolean vector on len( # samples ), with True for majority samples that are Tomek links.
- set_params(**params)[source]#
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.