Note
Go to the end to download the full example code.
Illustration of the definition of a Tomek link#
This example illustrates what is a Tomek link.
# Authors: Guillaume Lemaitre <g.lemaitre58@gmail.com>
# License: MIT
print(__doc__)
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_context("poster")
This function allows to make nice plotting
def make_plot_despine(ax):
sns.despine(ax=ax, offset=10)
ax.set_xlim([0, 3])
ax.set_ylim([0, 3])
ax.set_xlabel(r"$X_1$")
ax.set_ylabel(r"$X_2$")
ax.legend(loc="lower right")
We will generate some toy data that illustrates how
TomekLinks is used to clean a dataset.
import numpy as np
rng = np.random.RandomState(18)
X_minority = np.transpose(
[[1.1, 1.3, 1.15, 0.8, 0.55, 2.1], [1.0, 1.5, 1.7, 2.5, 0.55, 1.9]]
)
X_majority = np.transpose(
[
[2.1, 2.12, 2.13, 2.14, 2.2, 2.3, 2.5, 2.45],
[1.5, 2.1, 2.7, 0.9, 1.0, 1.4, 2.4, 2.9],
]
)
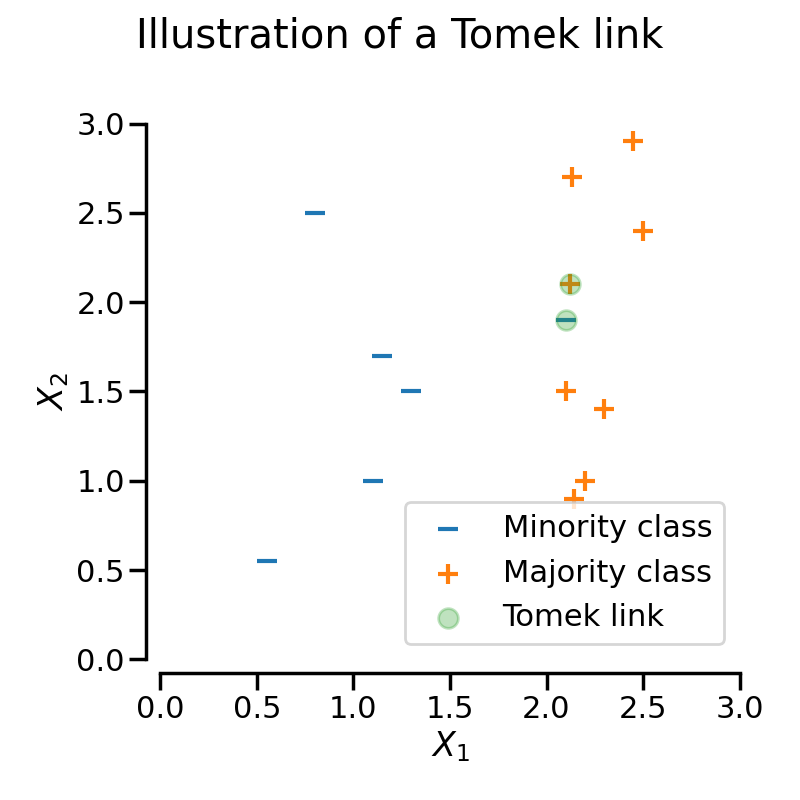
In the figure above, the samples highlighted in green form a Tomek link since they are of different classes and are nearest neighbors of each other.
fig, ax = plt.subplots(figsize=(8, 8))
ax.scatter(
X_minority[:, 0],
X_minority[:, 1],
label="Minority class",
s=200,
marker="_",
)
ax.scatter(
X_majority[:, 0],
X_majority[:, 1],
label="Majority class",
s=200,
marker="+",
)
# highlight the samples of interest
ax.scatter(
[X_minority[-1, 0], X_majority[1, 0]],
[X_minority[-1, 1], X_majority[1, 1]],
label="Tomek link",
s=200,
alpha=0.3,
)
make_plot_despine(ax)
fig.suptitle("Illustration of a Tomek link")
fig.tight_layout()
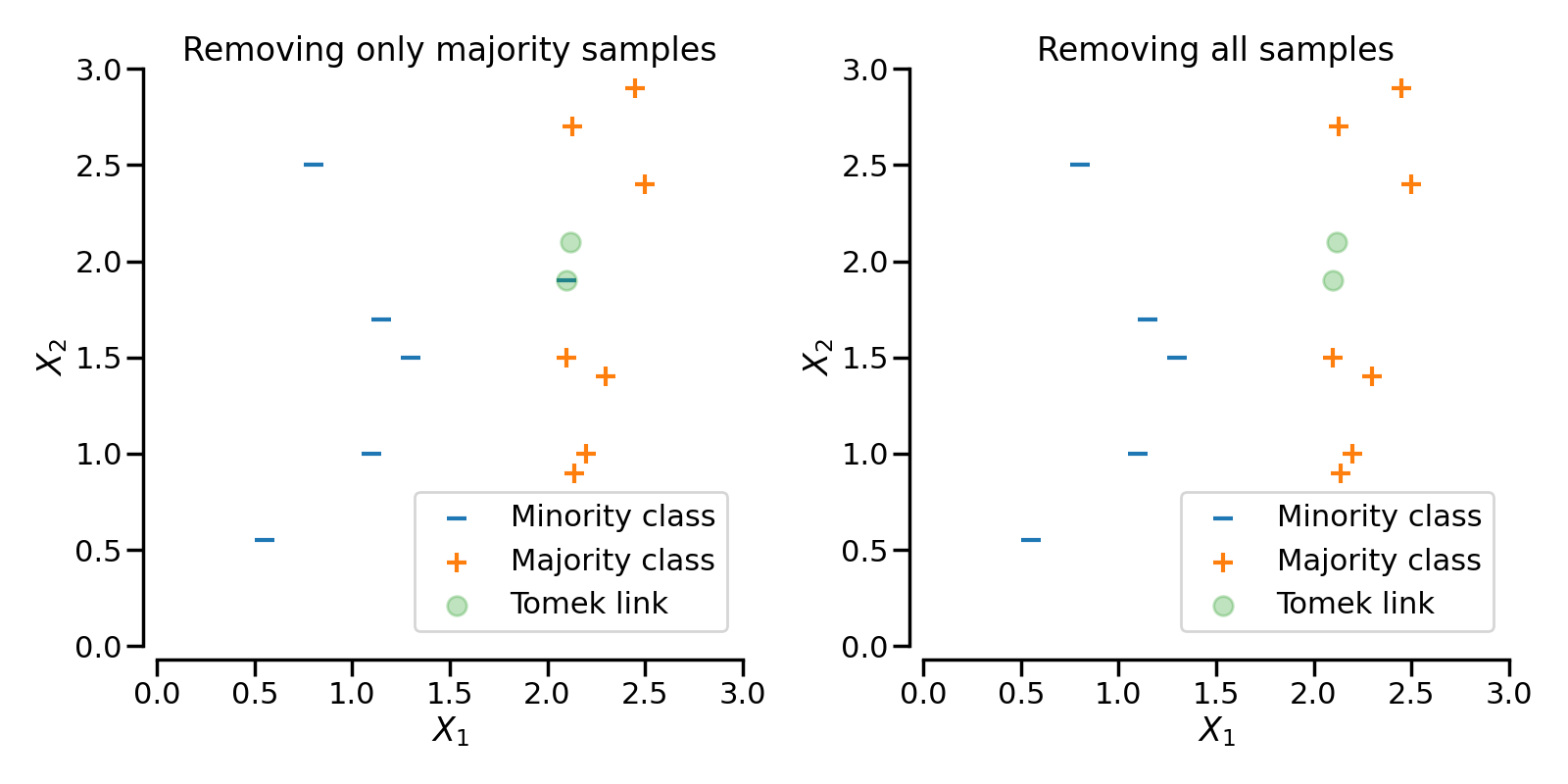
We can run the TomekLinks sampling to
remove the corresponding samples. If sampling_strategy='auto' only the
sample from the majority class will be removed. If sampling_strategy='all'
both samples will be removed.
from imblearn.under_sampling import TomekLinks
fig, axs = plt.subplots(nrows=1, ncols=2, figsize=(16, 8))
samplers = {
"Removing only majority samples": TomekLinks(sampling_strategy="auto"),
"Removing all samples": TomekLinks(sampling_strategy="all"),
}
for ax, (title, sampler) in zip(axs, samplers.items()):
X_res, y_res = sampler.fit_resample(
np.vstack((X_minority, X_majority)),
np.array([0] * X_minority.shape[0] + [1] * X_majority.shape[0]),
)
ax.scatter(
X_res[y_res == 0][:, 0],
X_res[y_res == 0][:, 1],
label="Minority class",
s=200,
marker="_",
)
ax.scatter(
X_res[y_res == 1][:, 0],
X_res[y_res == 1][:, 1],
label="Majority class",
s=200,
marker="+",
)
# highlight the samples of interest
ax.scatter(
[X_minority[-1, 0], X_majority[1, 0]],
[X_minority[-1, 1], X_majority[1, 1]],
label="Tomek link",
s=200,
alpha=0.3,
)
ax.set_title(title)
make_plot_despine(ax)
fig.tight_layout()
plt.show()
Total running time of the script: (0 minutes 2.069 seconds)
Estimated memory usage: 202 MB

