Note
Go to the end to download the full example code.
Customized sampler to implement an outlier rejections estimator#
This example illustrates the use of a custom sampler to implement an outlier rejections estimator. It can be used easily within a pipeline in which the number of samples can vary during training, which usually is a limitation of the current scikit-learn pipeline.
# Authors: Guillaume Lemaitre <g.lemaitre58@gmail.com>
# License: MIT
import matplotlib.pyplot as plt
import numpy as np
from sklearn.datasets import make_blobs, make_moons
from sklearn.ensemble import IsolationForest
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report
from imblearn import FunctionSampler
from imblearn.pipeline import make_pipeline
print(__doc__)
rng = np.random.RandomState(42)
def plot_scatter(X, y, title):
"""Function to plot some data as a scatter plot."""
plt.figure()
plt.scatter(X[y == 1, 0], X[y == 1, 1], label="Class #1")
plt.scatter(X[y == 0, 0], X[y == 0, 1], label="Class #0")
plt.legend()
plt.title(title)
Toy data generation#
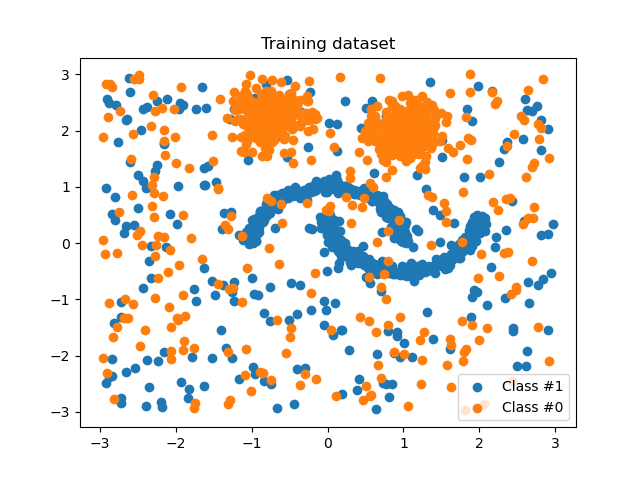
We are generating some non Gaussian data set contaminated with some unform noise.
moons, _ = make_moons(n_samples=500, noise=0.05)
blobs, _ = make_blobs(
n_samples=500, centers=[(-0.75, 2.25), (1.0, 2.0)], cluster_std=0.25
)
outliers = rng.uniform(low=-3, high=3, size=(500, 2))
X_train = np.vstack([moons, blobs, outliers])
y_train = np.hstack(
[
np.ones(moons.shape[0], dtype=np.int8),
np.zeros(blobs.shape[0], dtype=np.int8),
rng.randint(0, 2, size=outliers.shape[0], dtype=np.int8),
]
)
plot_scatter(X_train, y_train, "Training dataset")
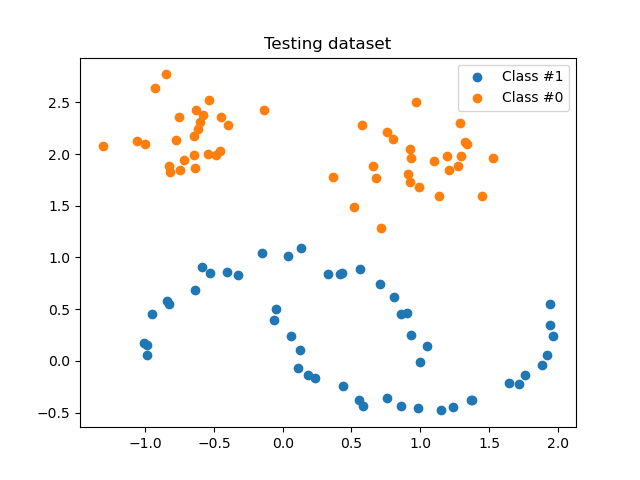
We will generate some cleaned test data without outliers.
moons, _ = make_moons(n_samples=50, noise=0.05)
blobs, _ = make_blobs(
n_samples=50, centers=[(-0.75, 2.25), (1.0, 2.0)], cluster_std=0.25
)
X_test = np.vstack([moons, blobs])
y_test = np.hstack(
[np.ones(moons.shape[0], dtype=np.int8), np.zeros(blobs.shape[0], dtype=np.int8)]
)
plot_scatter(X_test, y_test, "Testing dataset")
How to use the FunctionSampler#
We first define a function which will use
IsolationForest to eliminate some outliers from
our dataset during training. The function passed to the
FunctionSampler will be called when using the method
fit_resample.
def outlier_rejection(X, y):
"""This will be our function used to resample our dataset."""
model = IsolationForest(max_samples=100, contamination=0.4, random_state=rng)
model.fit(X)
y_pred = model.predict(X)
return X[y_pred == 1], y[y_pred == 1]
reject_sampler = FunctionSampler(func=outlier_rejection)
X_inliers, y_inliers = reject_sampler.fit_resample(X_train, y_train)
plot_scatter(X_inliers, y_inliers, "Training data without outliers")
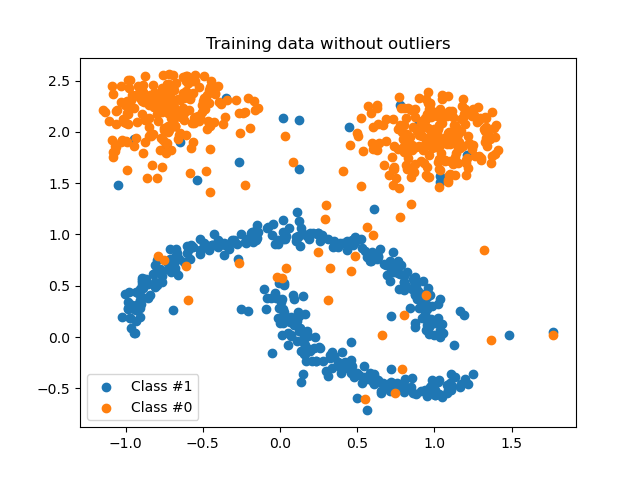
Integrate it within a pipeline#
By elimnating outliers before the training, the classifier will be less affected during the prediction.
pipe = make_pipeline(
FunctionSampler(func=outlier_rejection),
LogisticRegression(random_state=rng),
)
y_pred = pipe.fit(X_train, y_train).predict(X_test)
print(classification_report(y_test, y_pred))
clf = LogisticRegression(random_state=rng)
y_pred = clf.fit(X_train, y_train).predict(X_test)
print(classification_report(y_test, y_pred))
plt.show()
precision recall f1-score support
0 1.00 1.00 1.00 50
1 1.00 1.00 1.00 50
accuracy 1.00 100
macro avg 1.00 1.00 1.00 100
weighted avg 1.00 1.00 1.00 100
precision recall f1-score support
0 0.85 1.00 0.92 50
1 1.00 0.82 0.90 50
accuracy 0.91 100
macro avg 0.92 0.91 0.91 100
weighted avg 0.92 0.91 0.91 100
Total running time of the script: (0 minutes 1.789 seconds)
Estimated memory usage: 203 MB

