Note
Go to the end to download the full example code.
Compare sampler combining over- and under-sampling#
This example shows the effect of applying an under-sampling algorithms after SMOTE over-sampling. In the literature, Tomek’s link and edited nearest neighbours are the two methods which have been used and are available in imbalanced-learn.
# Authors: Guillaume Lemaitre <g.lemaitre58@gmail.com>
# License: MIT
print(__doc__)
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_context("poster")
Dataset generation#
We will create an imbalanced dataset with a couple of samples. We will use
make_classification to generate this dataset.
_, ax = plt.subplots(figsize=(6, 6))
_ = ax.scatter(X[:, 0], X[:, 1], c=y, alpha=0.8, edgecolor="k")
The following function will be used to plot the sample space after resampling to illustrate the characteristic of an algorithm.
from collections import Counter
def plot_resampling(X, y, sampler, ax):
"""Plot the resampled dataset using the sampler."""
X_res, y_res = sampler.fit_resample(X, y)
ax.scatter(X_res[:, 0], X_res[:, 1], c=y_res, alpha=0.8, edgecolor="k")
sns.despine(ax=ax, offset=10)
ax.set_title(f"Decision function for {sampler.__class__.__name__}")
return Counter(y_res)
The following function will be used to plot the decision function of a classifier given some data.
import numpy as np
def plot_decision_function(X, y, clf, ax):
"""Plot the decision function of the classifier and the original data"""
plot_step = 0.02
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(
np.arange(x_min, x_max, plot_step), np.arange(y_min, y_max, plot_step)
)
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
ax.contourf(xx, yy, Z, alpha=0.4)
ax.scatter(X[:, 0], X[:, 1], alpha=0.8, c=y, edgecolor="k")
ax.set_title(f"Resampling using {clf[0].__class__.__name__}")
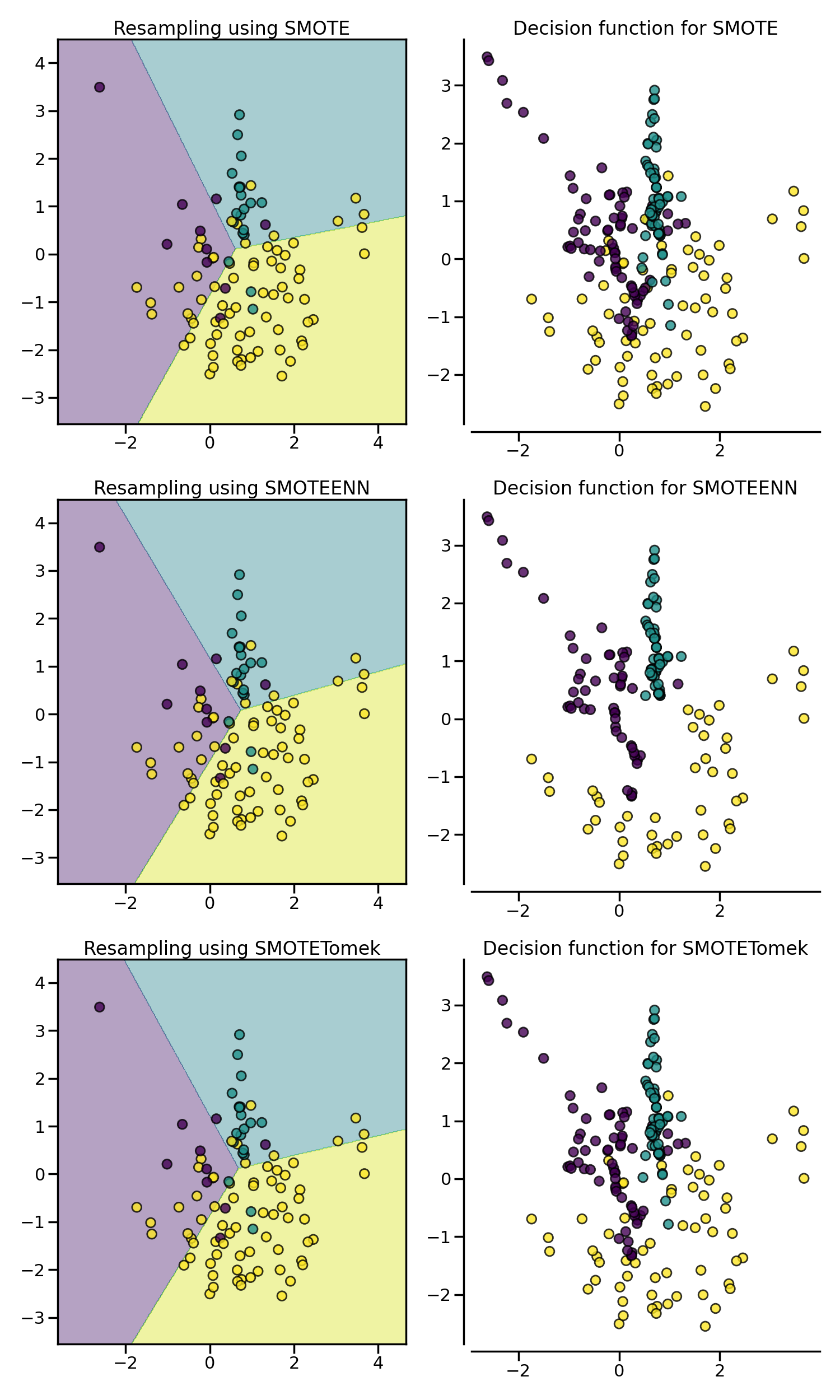
SMOTE allows to generate samples. However,
this method of over-sampling does not have any knowledge regarding the
underlying distribution. Therefore, some noisy samples can be generated, e.g.
when the different classes cannot be well separated. Hence, it can be
beneficial to apply an under-sampling algorithm to clean the noisy samples.
Two methods are usually used in the literature: (i) Tomek’s link and (ii)
edited nearest neighbours cleaning methods. Imbalanced-learn provides two
ready-to-use samplers SMOTETomek and
SMOTEENN. In general,
SMOTEENN cleans more noisy data than
SMOTETomek.
from sklearn.linear_model import LogisticRegression
from imblearn.combine import SMOTEENN, SMOTETomek
from imblearn.over_sampling import SMOTE
from imblearn.pipeline import make_pipeline
samplers = [SMOTE(random_state=0), SMOTEENN(random_state=0), SMOTETomek(random_state=0)]
fig, axs = plt.subplots(3, 2, figsize=(15, 25))
for ax, sampler in zip(axs, samplers):
clf = make_pipeline(sampler, LogisticRegression()).fit(X, y)
plot_decision_function(X, y, clf, ax[0])
plot_resampling(X, y, sampler, ax[1])
fig.tight_layout()
plt.show()
Total running time of the script: (0 minutes 2.023 seconds)
Estimated memory usage: 214 MB

